pypmc
pypmc is a python package focusing on adaptive importance sampling. It can be used for integration and sampling from a user-defined target density. A typical application is Bayesian inference, where one wants to sample from the posterior to marginalize over parameters and to compute the evidence. The key idea is to create a good proposal density by adapting a mixture of Gaussian or student's t components to the target density. The package is able to efficiently integrate multimodal functions in up to about 30-40 dimensions at the level of 1% accuracy or less. For many problems, this is achieved without requiring any manual input from the user about details of the function. pypmc supports importance sampling on a cluster of machines via mpi4py out of the box.
Useful tools that can be used stand-alone include:
- importance sampling (sampling & integration)
- adaptive Markov chain Monte Carlo (sampling)
- variational Bayes (clustering)
- population Monte Carlo (clustering)
Installation
Instructions are maintained here.
Getting started
Fully documented examples are shipped in the examples subdirectory of the source distribution or available online including sample output here. Feel free to save and modify them according to your needs.
Documentation
The full documentation with a manual and api description is available at here.
Credits
pypmc was developed by Stephan Jahn (TU Munich) under the supervision of Frederik Beaujean (LMU Munich) as part of Stephan's master's thesis at the Excellence Cluster Universe, Garching, Germany, in 2014.
If you use pypmc in academic work, we kindly ask you to cite the respective release as indicated by the zenodo DOI above. Thanks!
Day to day maintenance is assisted by Danny van Dyk.
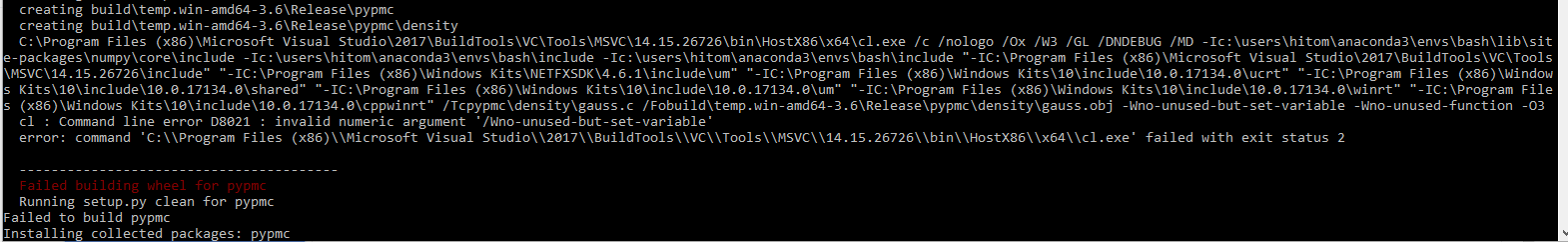 I went into the setup.py and fiddled with the code and found that these lines were causing the errors:
I went into the setup.py and fiddled with the code and found that these lines were causing the errors:
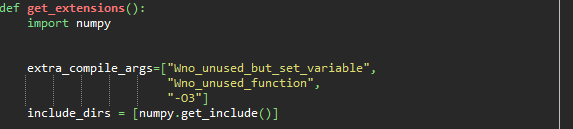 It allows me to install pypmc if I delete the commands inside the extra_compile_args function, but then it doesn't function properly as it can not import the name 'gauss'.
It allows me to install pypmc if I delete the commands inside the extra_compile_args function, but then it doesn't function properly as it can not import the name 'gauss'.